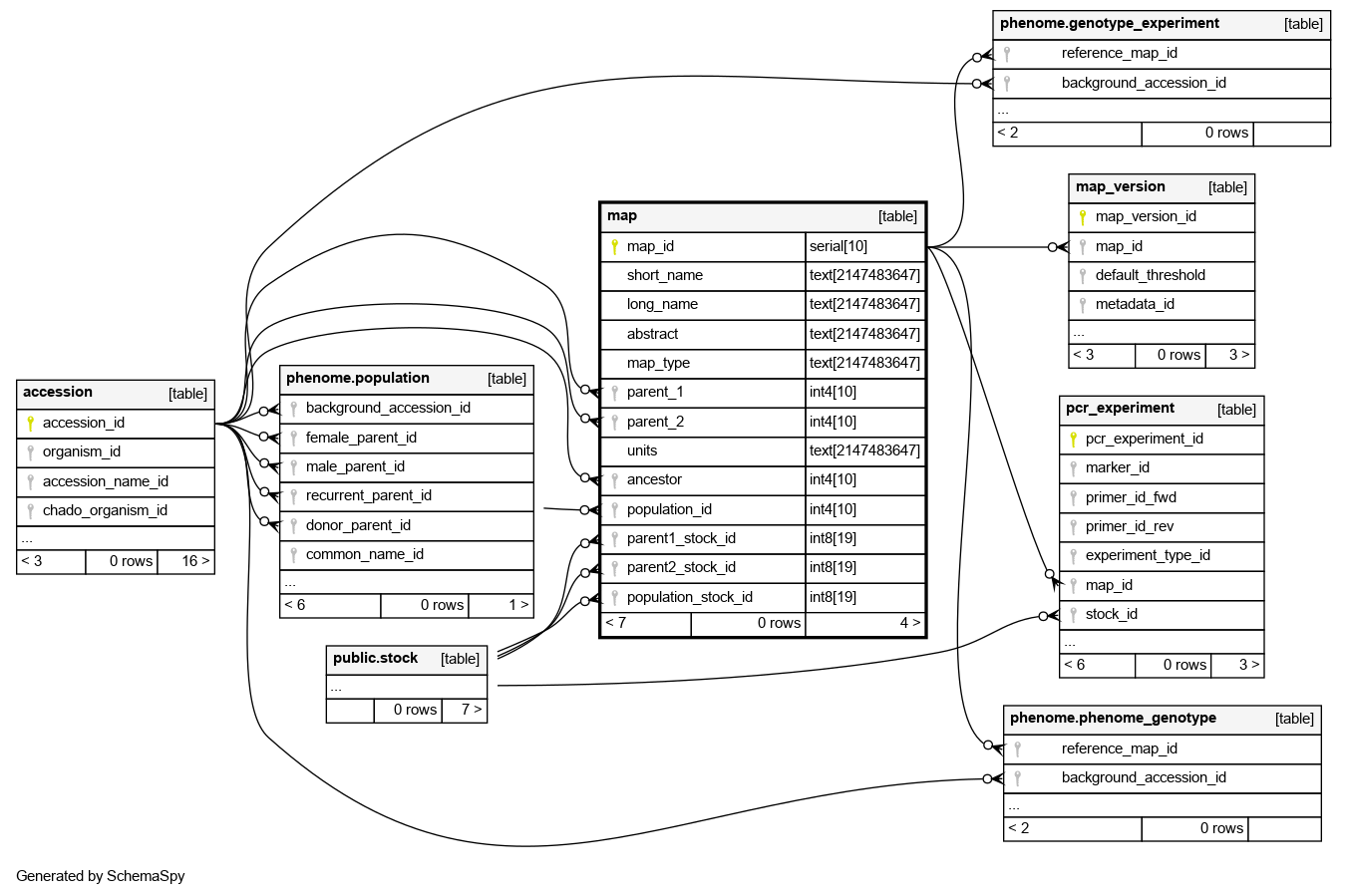
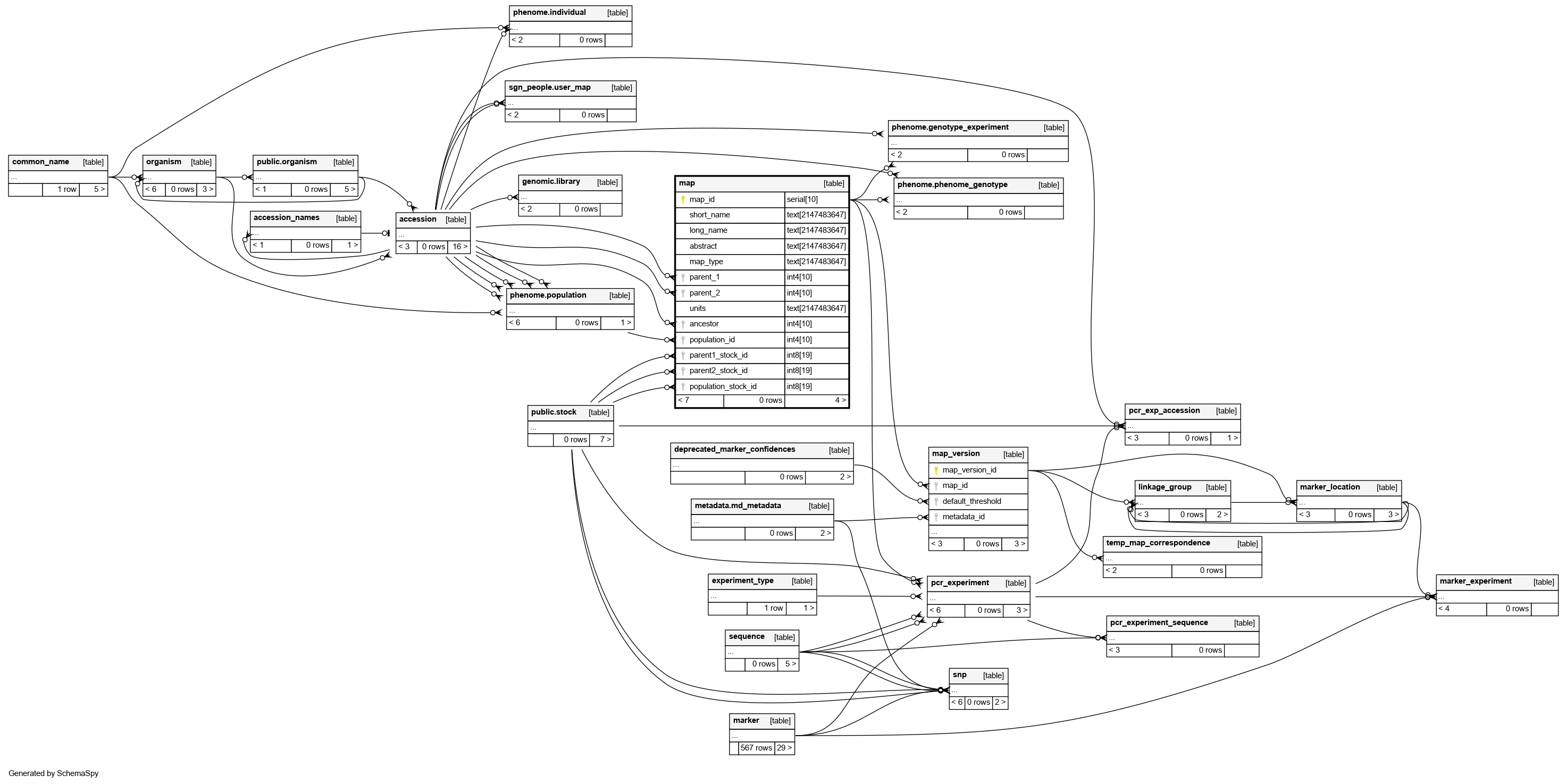
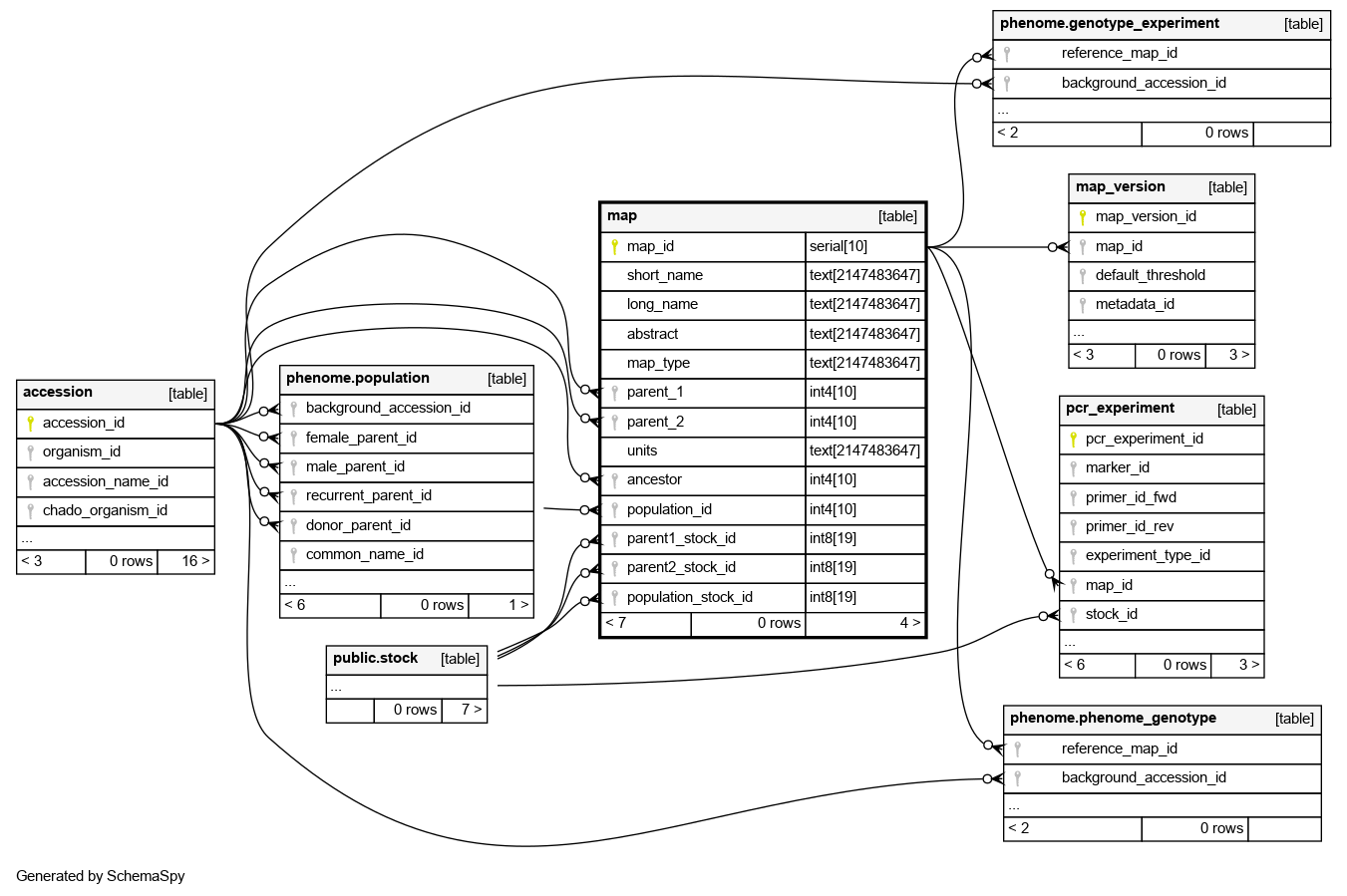
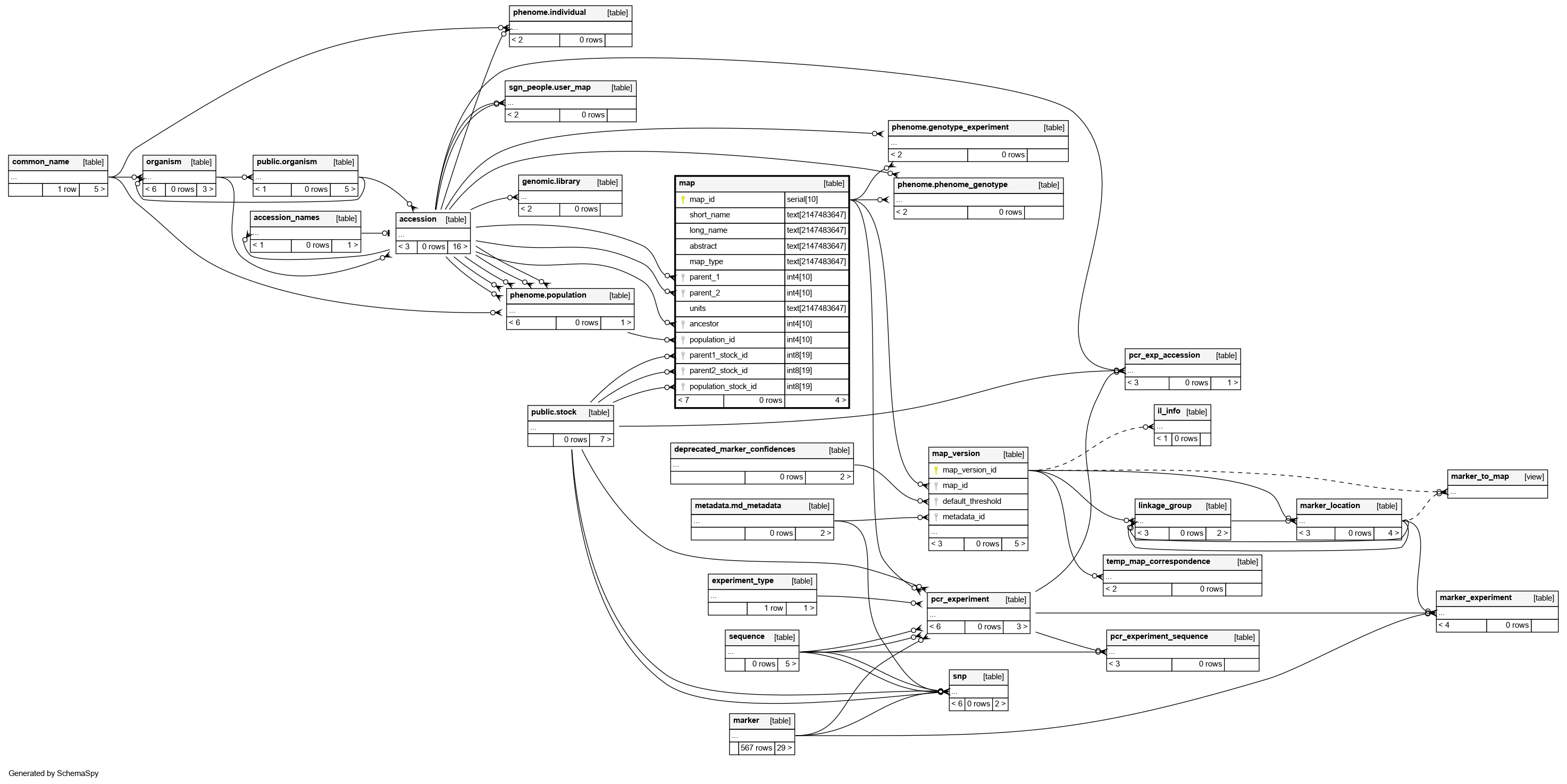
Columns
| Column | Type | Size | Nulls | Auto | Default | Children | Parents | Comments | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| map_id | serial | 10 | √ | nextval('sgn.map_map_id_seq'::regclass) |
|
|
||||||||||||||
| short_name | text | 2147483647 | null |
|
|
|||||||||||||||
| long_name | text | 2147483647 | √ | null |
|
|
||||||||||||||
| abstract | text | 2147483647 | √ | null |
|
|
||||||||||||||
| map_type | text | 2147483647 | √ | null |
|
|
||||||||||||||
| parent_1 | int4 | 10 | √ | null |
|
|
||||||||||||||
| parent_2 | int4 | 10 | √ | null |
|
|
||||||||||||||
| units | text | 2147483647 | √ | 'cM'::text |
|
|
||||||||||||||
| ancestor | int4 | 10 | √ | null |
|
|
||||||||||||||
| population_id | int4 | 10 | √ | null |
|
|
||||||||||||||
| parent1_stock_id | int8 | 19 | √ | null |
|
|
||||||||||||||
| parent2_stock_id | int8 | 19 | √ | null |
|
|
||||||||||||||
| population_stock_id | int8 | 19 | √ | null |
|
|
Indexes
| Constraint Name | Type | Sort | Column(s) |
|---|---|---|---|
| map_pkey | Primary key | Asc | map_id |
Check Constraints
| Constraint Name | Constraint |
|---|---|
| map_units_check | (((units = 'cM'::text) OR (units = 'Mb'::text))) |
| map_map_type_check | (((map_type = 'genetic'::text) OR (map_type = 'fish'::text) OR (map_type = 'sequence'::text) OR (map_type = 'QTL'::text))) |