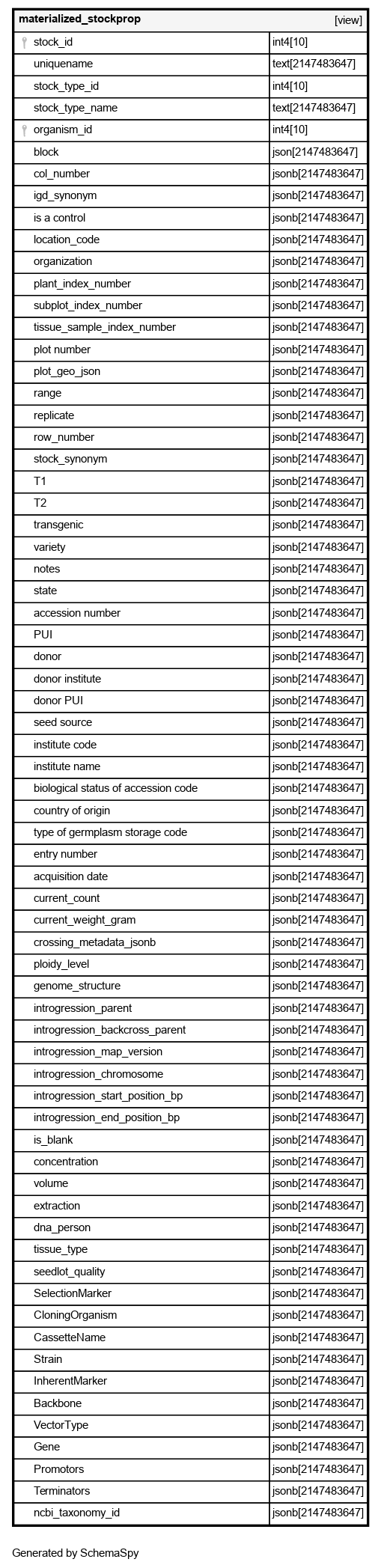
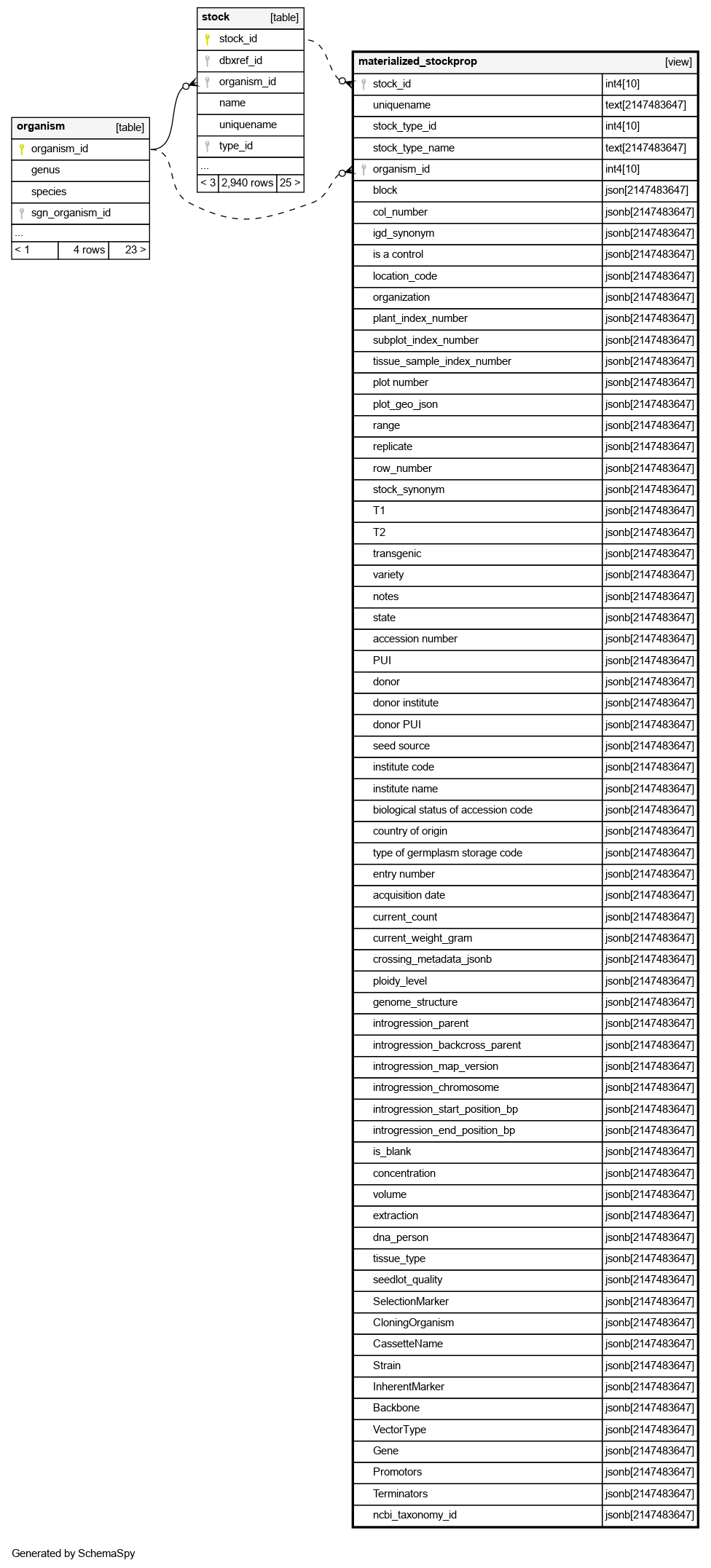
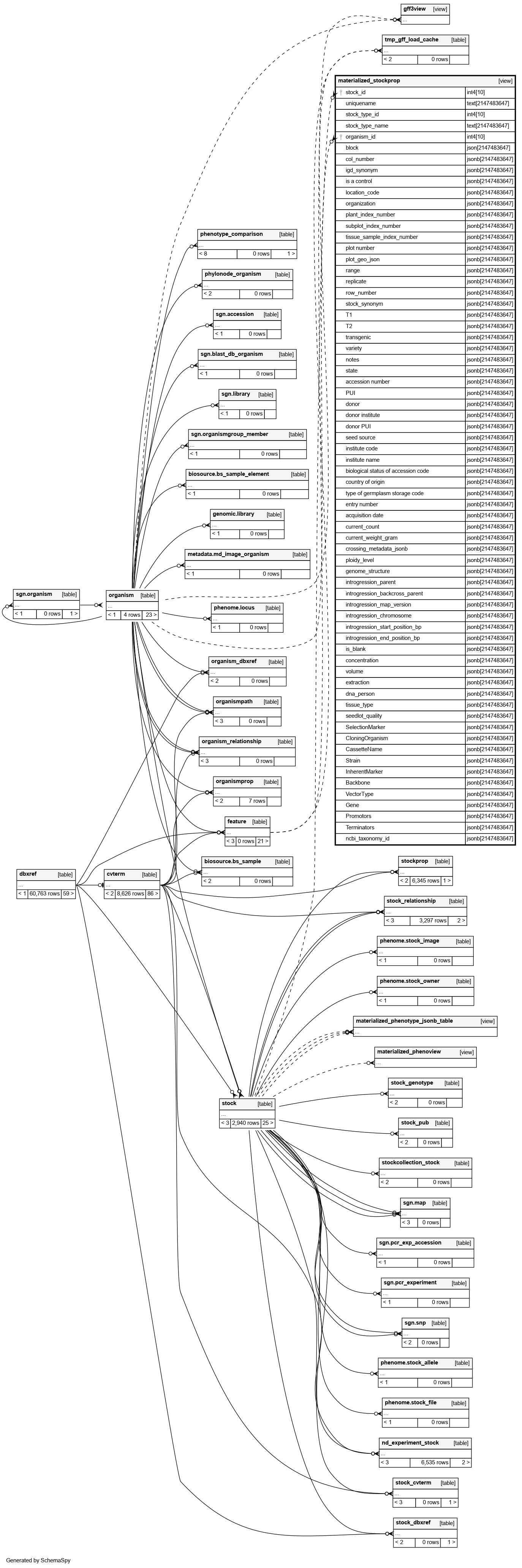
Columns
| Column | Type | Size | Nulls | Auto | Default | Children | Parents | Comments | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| stock_id | int4 | 10 | √ | null |
|
|
|||||
| uniquename | text | 2147483647 | √ | null |
|
|
|||||
| stock_type_id | int4 | 10 | √ | null |
|
|
|||||
| stock_type_name | text | 2147483647 | √ | null |
|
|
|||||
| organism_id | int4 | 10 | √ | null |
|
|
|||||
| block | json | 2147483647 | √ | null |
|
|
|||||
| col_number | jsonb | 2147483647 | √ | null |
|
|
|||||
| igd_synonym | jsonb | 2147483647 | √ | null |
|
|
|||||
| is a control | jsonb | 2147483647 | √ | null |
|
|
|||||
| location_code | jsonb | 2147483647 | √ | null |
|
|
|||||
| organization | jsonb | 2147483647 | √ | null |
|
|
|||||
| plant_index_number | jsonb | 2147483647 | √ | null |
|
|
|||||
| subplot_index_number | jsonb | 2147483647 | √ | null |
|
|
|||||
| tissue_sample_index_number | jsonb | 2147483647 | √ | null |
|
|
|||||
| plot number | jsonb | 2147483647 | √ | null |
|
|
|||||
| plot_geo_json | jsonb | 2147483647 | √ | null |
|
|
|||||
| range | jsonb | 2147483647 | √ | null |
|
|
|||||
| replicate | jsonb | 2147483647 | √ | null |
|
|
|||||
| row_number | jsonb | 2147483647 | √ | null |
|
|
|||||
| stock_synonym | jsonb | 2147483647 | √ | null |
|
|
|||||
| T1 | jsonb | 2147483647 | √ | null |
|
|
|||||
| T2 | jsonb | 2147483647 | √ | null |
|
|
|||||
| transgenic | jsonb | 2147483647 | √ | null |
|
|
|||||
| variety | jsonb | 2147483647 | √ | null |
|
|
|||||
| notes | jsonb | 2147483647 | √ | null |
|
|
|||||
| state | jsonb | 2147483647 | √ | null |
|
|
|||||
| accession number | jsonb | 2147483647 | √ | null |
|
|
|||||
| PUI | jsonb | 2147483647 | √ | null |
|
|
|||||
| donor | jsonb | 2147483647 | √ | null |
|
|
|||||
| donor institute | jsonb | 2147483647 | √ | null |
|
|
|||||
| donor PUI | jsonb | 2147483647 | √ | null |
|
|
|||||
| seed source | jsonb | 2147483647 | √ | null |
|
|
|||||
| institute code | jsonb | 2147483647 | √ | null |
|
|
|||||
| institute name | jsonb | 2147483647 | √ | null |
|
|
|||||
| biological status of accession code | jsonb | 2147483647 | √ | null |
|
|
|||||
| country of origin | jsonb | 2147483647 | √ | null |
|
|
|||||
| type of germplasm storage code | jsonb | 2147483647 | √ | null |
|
|
|||||
| entry number | jsonb | 2147483647 | √ | null |
|
|
|||||
| acquisition date | jsonb | 2147483647 | √ | null |
|
|
|||||
| current_count | jsonb | 2147483647 | √ | null |
|
|
|||||
| current_weight_gram | jsonb | 2147483647 | √ | null |
|
|
|||||
| crossing_metadata_jsonb | jsonb | 2147483647 | √ | null |
|
|
|||||
| ploidy_level | jsonb | 2147483647 | √ | null |
|
|
|||||
| genome_structure | jsonb | 2147483647 | √ | null |
|
|
|||||
| introgression_parent | jsonb | 2147483647 | √ | null |
|
|
|||||
| introgression_backcross_parent | jsonb | 2147483647 | √ | null |
|
|
|||||
| introgression_map_version | jsonb | 2147483647 | √ | null |
|
|
|||||
| introgression_chromosome | jsonb | 2147483647 | √ | null |
|
|
|||||
| introgression_start_position_bp | jsonb | 2147483647 | √ | null |
|
|
|||||
| introgression_end_position_bp | jsonb | 2147483647 | √ | null |
|
|
|||||
| is_blank | jsonb | 2147483647 | √ | null |
|
|
|||||
| concentration | jsonb | 2147483647 | √ | null |
|
|
|||||
| volume | jsonb | 2147483647 | √ | null |
|
|
|||||
| extraction | jsonb | 2147483647 | √ | null |
|
|
|||||
| dna_person | jsonb | 2147483647 | √ | null |
|
|
|||||
| tissue_type | jsonb | 2147483647 | √ | null |
|
|
|||||
| seedlot_quality | jsonb | 2147483647 | √ | null |
|
|
|||||
| SelectionMarker | jsonb | 2147483647 | √ | null |
|
|
|||||
| CloningOrganism | jsonb | 2147483647 | √ | null |
|
|
|||||
| CassetteName | jsonb | 2147483647 | √ | null |
|
|
|||||
| Strain | jsonb | 2147483647 | √ | null |
|
|
|||||
| InherentMarker | jsonb | 2147483647 | √ | null |
|
|
|||||
| Backbone | jsonb | 2147483647 | √ | null |
|
|
|||||
| VectorType | jsonb | 2147483647 | √ | null |
|
|
|||||
| Gene | jsonb | 2147483647 | √ | null |
|
|
|||||
| Promotors | jsonb | 2147483647 | √ | null |
|
|
|||||
| Terminators | jsonb | 2147483647 | √ | null |
|
|
|||||
| ncbi_taxonomy_id | jsonb | 2147483647 | √ | null |
|
|