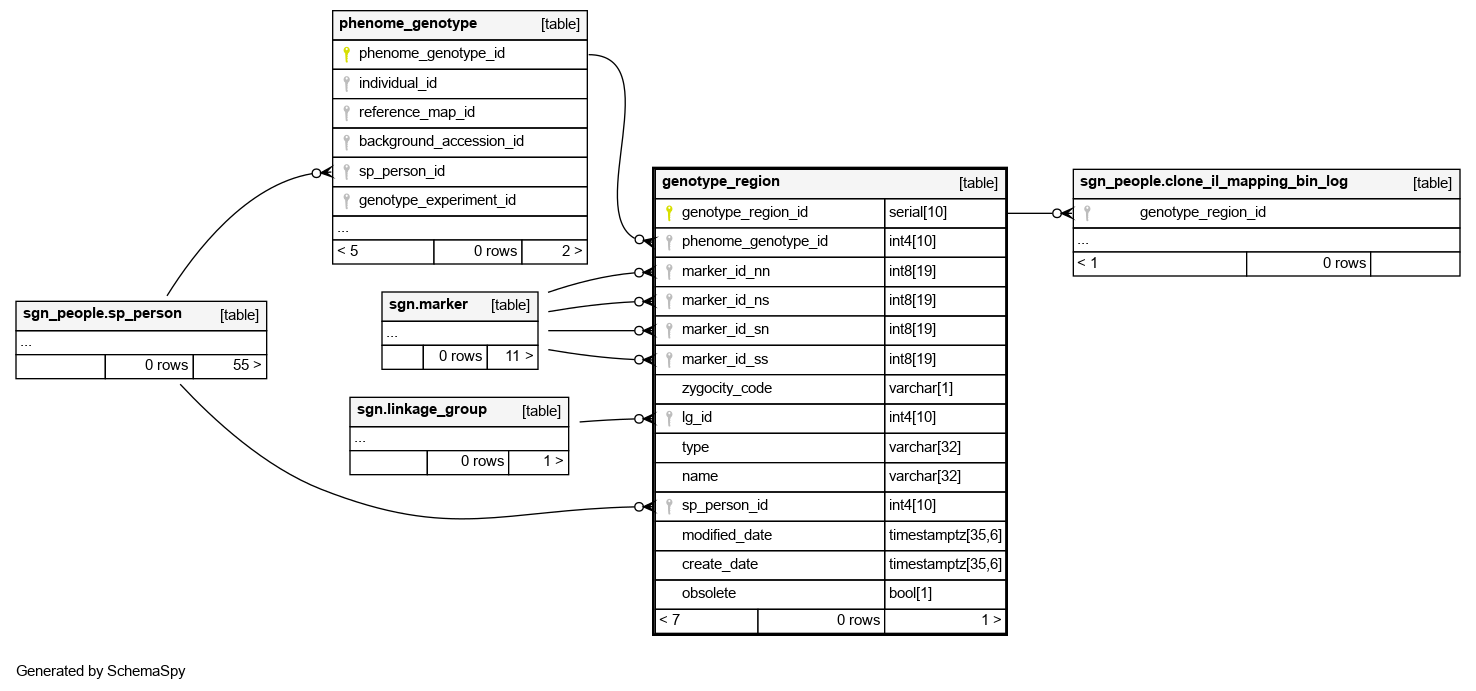
Columns
| Column | Type | Size | Nulls | Auto | Default | Children | Parents | Comments | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| genotype_region_id | serial | 10 | √ | nextval('phenome.genotype_region_genotype_region_id_seq'::regclass) |
|
|
|||||
| phenome_genotype_id | int4 | 10 | √ | null |
|
|
optional genotype this region belongs to. some regions are artificial, arising from combinations of other regions, and thus do not have an associated genotype |
||||
| marker_id_nn | int8 | 19 | √ | null |
|
|
the north marker in the pair of markers bracketing the north end of this region. this may be null for regions at the north end of a linkage group |
||||
| marker_id_ns | int8 | 19 | null |
|
|
the south marker in the pair of markers bracketing the north end of this region |
|||||
| marker_id_sn | int8 | 19 | null |
|
|
the north marker in the pair of markers bracketing the south end of this region |
|||||
| marker_id_ss | int8 | 19 | √ | null |
|
|
the south marker in the pair of markers bracketing the south end of this region. this may be null for regions at the south end of a linkage group. |
||||
| zygocity_code | varchar | 1 | √ | null |
|
|
|||||
| lg_id | int4 | 10 | √ | null |
|
|
the linkage group in a specific version of a specific map where this region is located |
||||
| type | varchar | 32 | null |
|
|
the type of polymorphic region this is. map is mapping experiment data, inbred is IL lines segments, and bin is a derived region based on a boolean combination of inbred fragments. For bin regions, the specific boolean combination of fragments that make the bin is not stored. |
|||||
| name | varchar | 32 | √ | null |
|
|
special name for this region, if any. optional |
||||
| sp_person_id | int4 | 10 | √ | null |
|
|
the person who loaded this datum. optional |
||||
| modified_date | timestamptz | 35,6 | now() |
|
|
||||||
| create_date | timestamptz | 35,6 | now() |
|
|
||||||
| obsolete | bool | 1 | false |
|
|
Indexes
| Constraint Name | Type | Sort | Column(s) |
|---|---|---|---|
| genotype_region_pkey | Primary key | Asc | genotype_region_id |
| phenome_genotype_marker_id_nn_idx | Performance | Asc | marker_id_nn |
| phenome_genotype_marker_id_ns_idx | Performance | Asc | marker_id_ns |
| phenome_genotype_marker_id_sn_idx | Performance | Asc | marker_id_sn |
| phenome_genotype_marker_id_ss_idx | Performance | Asc | marker_id_ss |
| phenome_genotype_region_genotype_id_idx | Performance | Asc | phenome_genotype_id |
| phenome_genotype_region_name | Performance | Asc | name |
| phenome_genotype_region_type | Performance | Asc | type |
| phenome_genotype_region_zygocity_code | Performance | Asc | zygocity_code |
Check Constraints
| Constraint Name | Constraint |
|---|---|
| genotype_region_zygocity_code_check | ((((zygocity_code)::text = 'a'::text) OR ((zygocity_code)::text = 'b'::text) OR ((zygocity_code)::text = 'c'::text) OR ((zygocity_code)::text = 'd'::text) OR ((zygocity_code)::text = 'h'::text))) |
| genotype_region_type_check | ((((type)::text = 'bin'::text) OR ((type)::text = 'map'::text) OR ((type)::text = 'inbred'::text))) |